Visualizing Survival Data with the {ggsurvfit} R Package
{ggsurvfit}
Licensing
This work is licensed under Creative Commons Zero v1.0 Universal.
Authors
Daniel D. Sjoberg


Senior Principal Data Scientist at Genentech.
Previously, a Lead Data Science Manager at the Prostate Cancer Clinical Trials Consortium, and a Senior Biostatistician at Memorial Sloan Kettering Cancer Center in New York City.
He enjoys R package development, creating many packages available on CRAN, R-Universe, and GitHub.
Winner of the 2021 American Statistical Association (ASA) Innovation in Statistical Programming and Analytics award.
Outline
What is survival analysis?
Visualizing Kaplan-Meier
Visualizing Competing Risks
Advanced Topics in {ggsurvfit}
- Combining figures
- How Risk Tables are constructed
- CDISC Helpers
Survival Analysis
Clark, T., Bradburn, M., Love, S., & Altman, D. (2003). Survival analysis part I: Basic concepts and first analyses. 232-238. ISSN 0007-0920.
M J Bradburn, T G Clark, S B Love, & D G Altman. (2003). _Survival Analysis Part II: Multivariate data analysis – an introduction to concepts and methods. British Journal of Cancer, 89(3), 431-436.
Bradburn, M., Clark, T., Love, S., & Altman, D. (2003). Survival analysis Part III: Multivariate data analysis – choosing a model and assessing its adequacy and fit_. 89(4), 605-11.
Clark, T., Bradburn, M., Love, S., & Altman, D. (2003). Survival analysis part IV: Further concepts and methods in survival analysis. 781-786. ISSN 0007-0920.
Survival Analysis
Survival times are data that measure follow-up time from a defined starting point to the occurrence of a given event, for example the time from the beginning to the end of a remission period or the time from the diagnosis of a disease to death. Standard statistical techniques cannot usually be applied because the data are often ‘censored’. A survival time is described as censored when there is a follow-up time but the event has not yet occurred.
Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care. 2004 Oct;8(5):389-94.
Survival Analysis
Because survival data are common in many fields, it also goes by other names.
Other Names
Reliability analysis
Duration analysis
Event history analysis
Time-to-event analysis
Examples
Time from surgery to death
Time from start of treatment to progression
Time from HIV infection to development of AIDS
Time to heart attack
Time to onset of substance abuse
Time to initiation of sexual activity
Time to machine malfunction
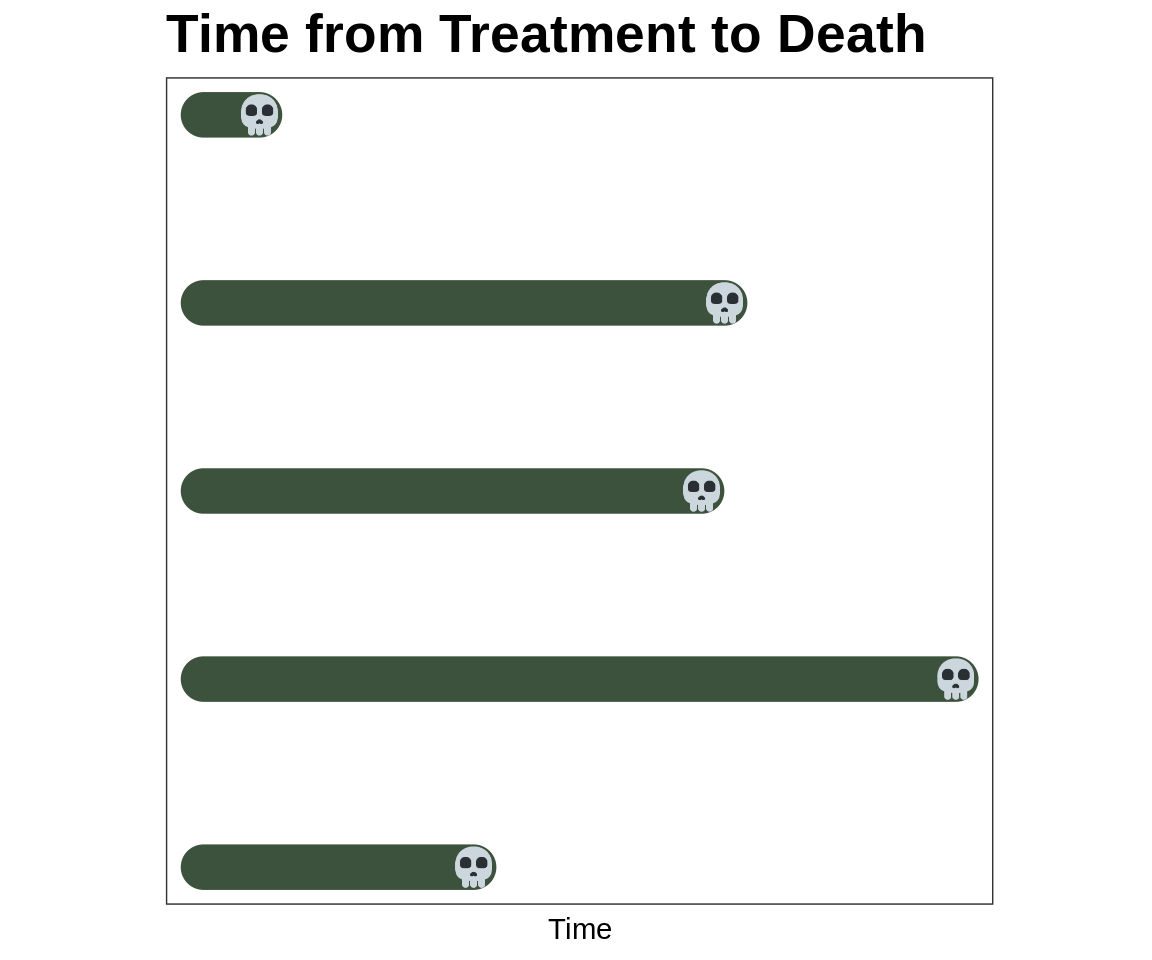
Survival Analysis, No Censoring
The skulls are the time when each of these patients die
We observe the follow-up in the solid line
How can we summarize the time from treatment to death?
Survival Analysis, Censoring
The skulls are the time when each of these patients die
We observe the follow-up in the solid line
The dashed line is unobserved
How can we summarize the average time from treatment to death?
Survival Analysis Methods
| Standard Method | Survival Method1 |
|---|---|
| Means/Median/Percentiles | Kaplan-Meier Estimator |
| t-test/Rank-sum Test | Log-rank Test |
| Linear/Logistic Regression | Cox Proportional Hazards Regression2 |
| 1 There are multiple methods available tailored to specific cases and these are the most common. | |
| 2 We will not be covering regression methods today. | |
Why {ggsurvfit} ?
Why {ggsurvfit} ?
Use ggplot2 functions
- Each
ggsurvfitfunction is written as a properggplot2geom - Package functions woven with ggplot2 functions seamlessly
- Don’t need to learn to style with
ggsurvfitfunctions - Use your ggplot2 knowledge if you want to customize
- Each
Limitless customization
- Modify x-axis scales or any other plot feature and risk table will still align with plot
Simple saving and export through
ggplot2::ggsave()Ready to publish legends; raw variable names do not appear in the figure
Data
The following examples use a labeled version of the survival::colon data set.
Data
library(ggsurvfit)
library(gtsummary)
library(tidyverse)
df_colon <- df_colon |> select(time, status, surg)
df_colon# A tibble: 929 × 3
time status surg
<dbl> <dbl> <fct>
1 2.65 1 Limited Time Since Surgery
2 8.45 0 Limited Time Since Surgery
3 1.48 1 Limited Time Since Surgery
4 0.671 1 Extended Time Since Surgery
5 1.43 1 Extended Time Since Surgery
6 2.48 1 Limited Time Since Surgery
7 0.627 1 Extended Time Since Surgery
8 8.74 0 Limited Time Since Surgery
9 8.69 0 Limited Time Since Surgery
10 9.06 0 Extended Time Since Surgery
# ℹ 919 more rowsData
| Characteristic | N = 929 |
|---|---|
| Follow-up time, years, Median (IQR) | 4.24 (1.01, 6.27) |
| Recurrence Status, n / N | 468 / 929 |
| Time from Surgery to Treatment, n (%) | |
| Limited Time Since Surgery | 682 (73%) |
| Extended Time Since Surgery | 247 (27%) |
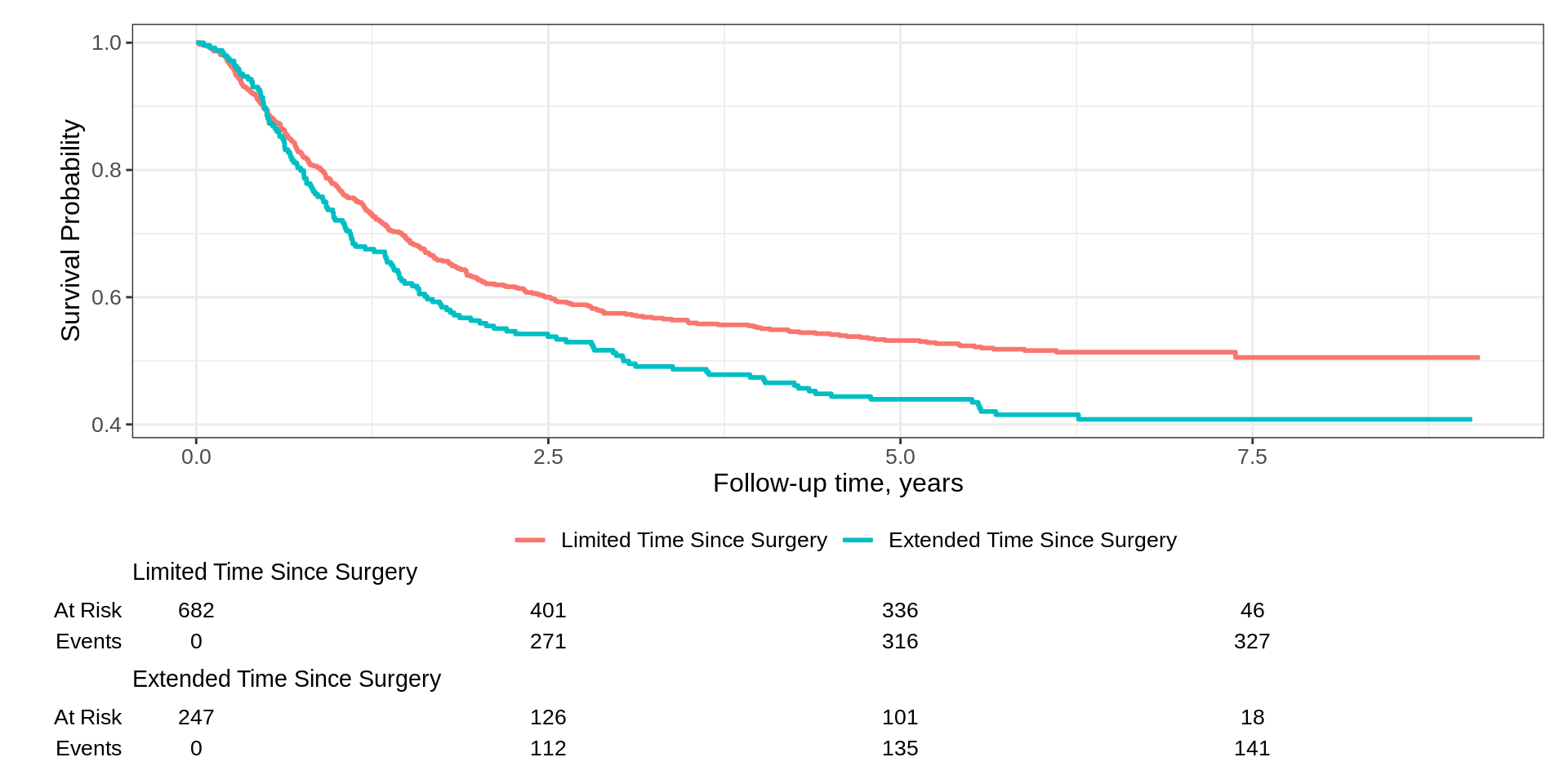
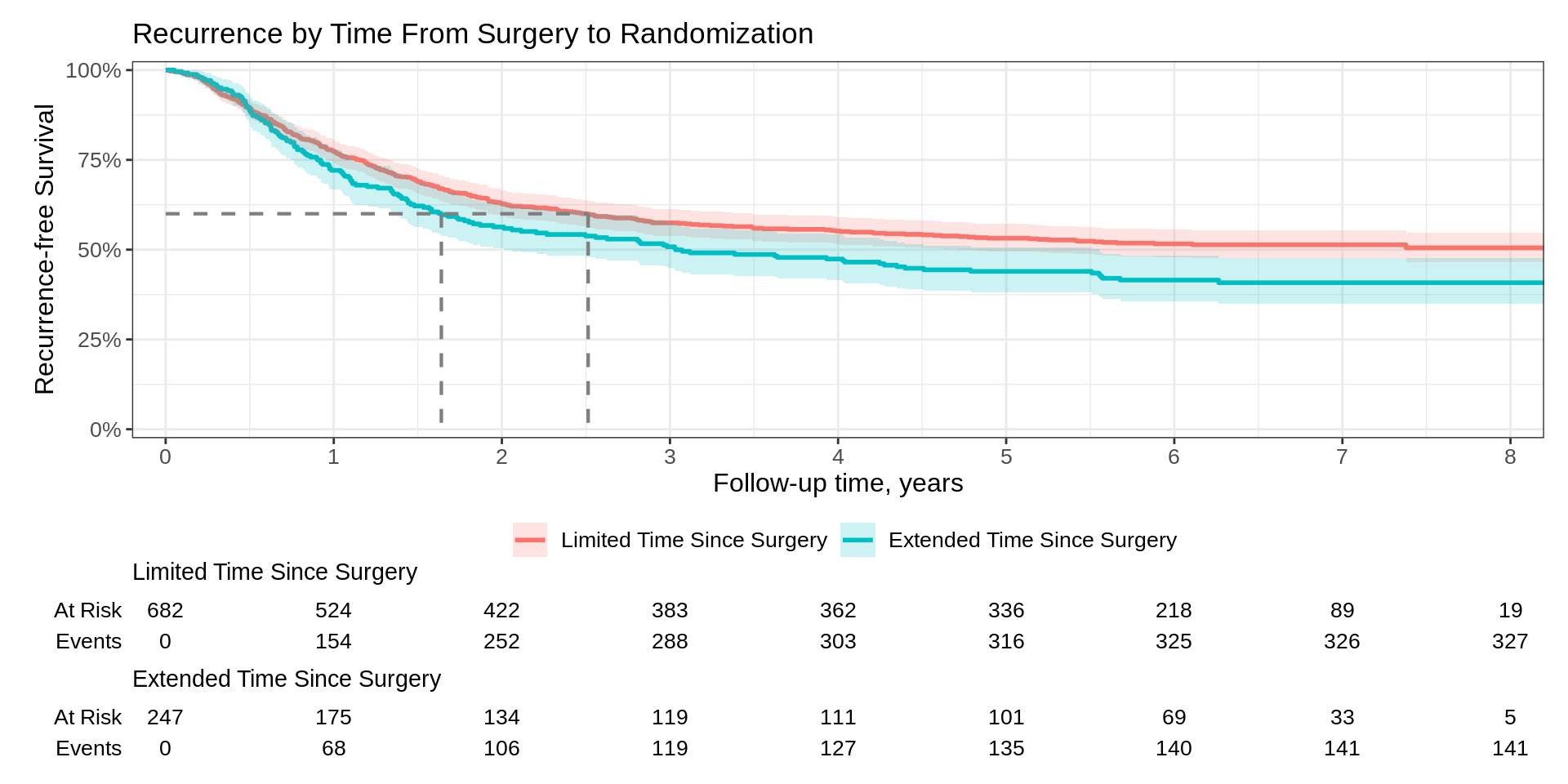
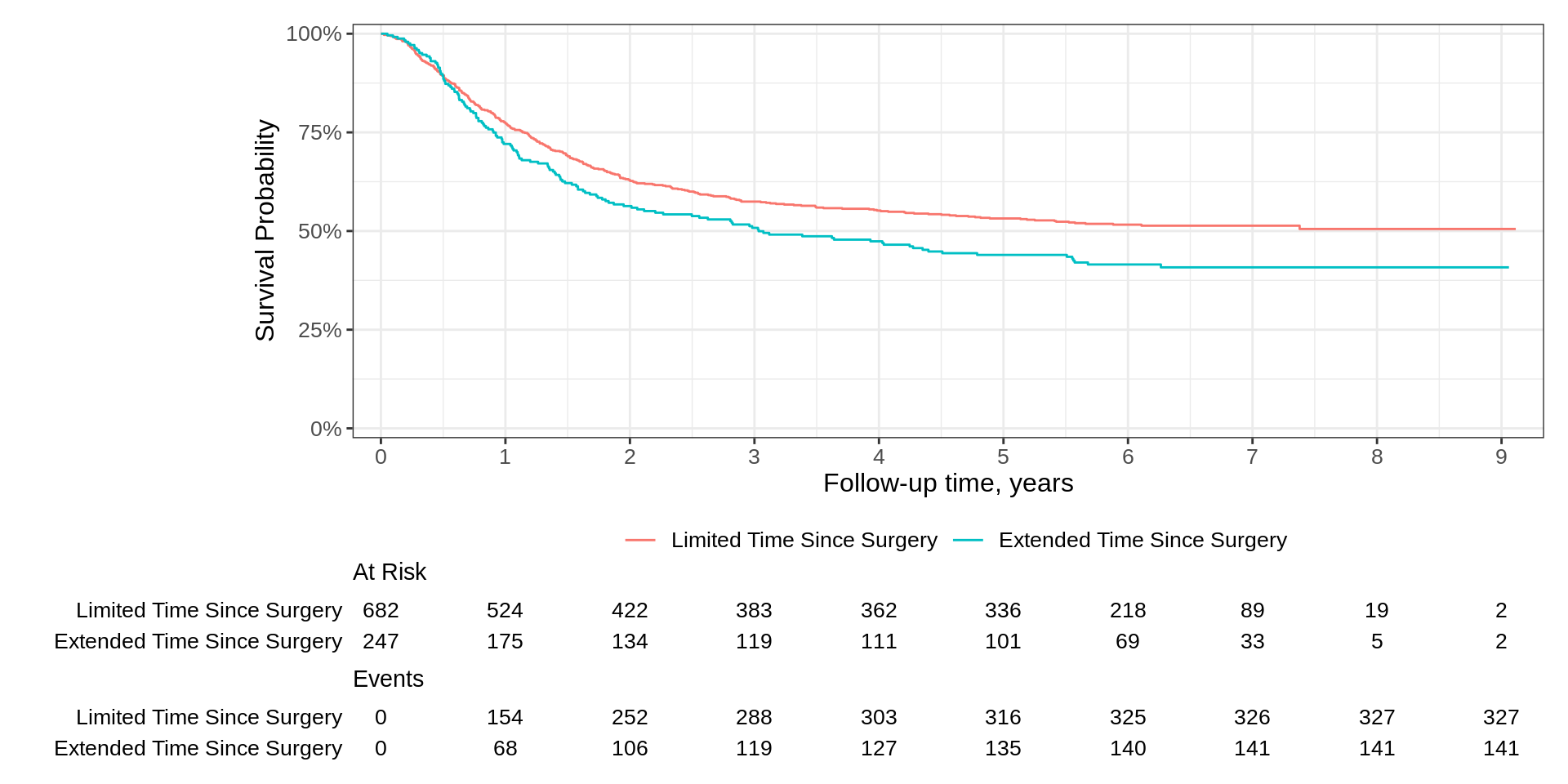
Kaplan-Meier Basics
How do we calculate the Kaplan-Meier estimator?
They look the same, right?
Call: survfit(formula = Surv(time, status) ~ surg, data = df_colon)
n events median 0.95LCL 0.95UCL
surg=Limited Time Since Surgery 682 327 NA 4.77 NA
surg=Extended Time Since Surgery 247 141 3.03 2.01 5.55Kaplan-Meier Basics
What is the difference?
ggsurvfit::survfit2()tracks the environment from which the call was madeAccurately reconstruct or parse call at any point post estimation
Call is parsed when p-values are reported and when labels are created
Most functions in the package work with both
survfit2()andsurvfit(); however, the output will be styled in a preferable format withsurvfit2().
Basic Example
- The Good
- Simple code and figure is nearly publishable
- Risk table with both no. at risk and events easily added
- x-axis label taken from the
timecolumn label - Can use ggplot2
+notation
- The Could-Be-Better
- y-axis label is incorrect, and the range of axis is best at 0-100%
- Axis padding a bit more than I prefer for a KM figure
- x-axis typically has more tick marks for KM figure
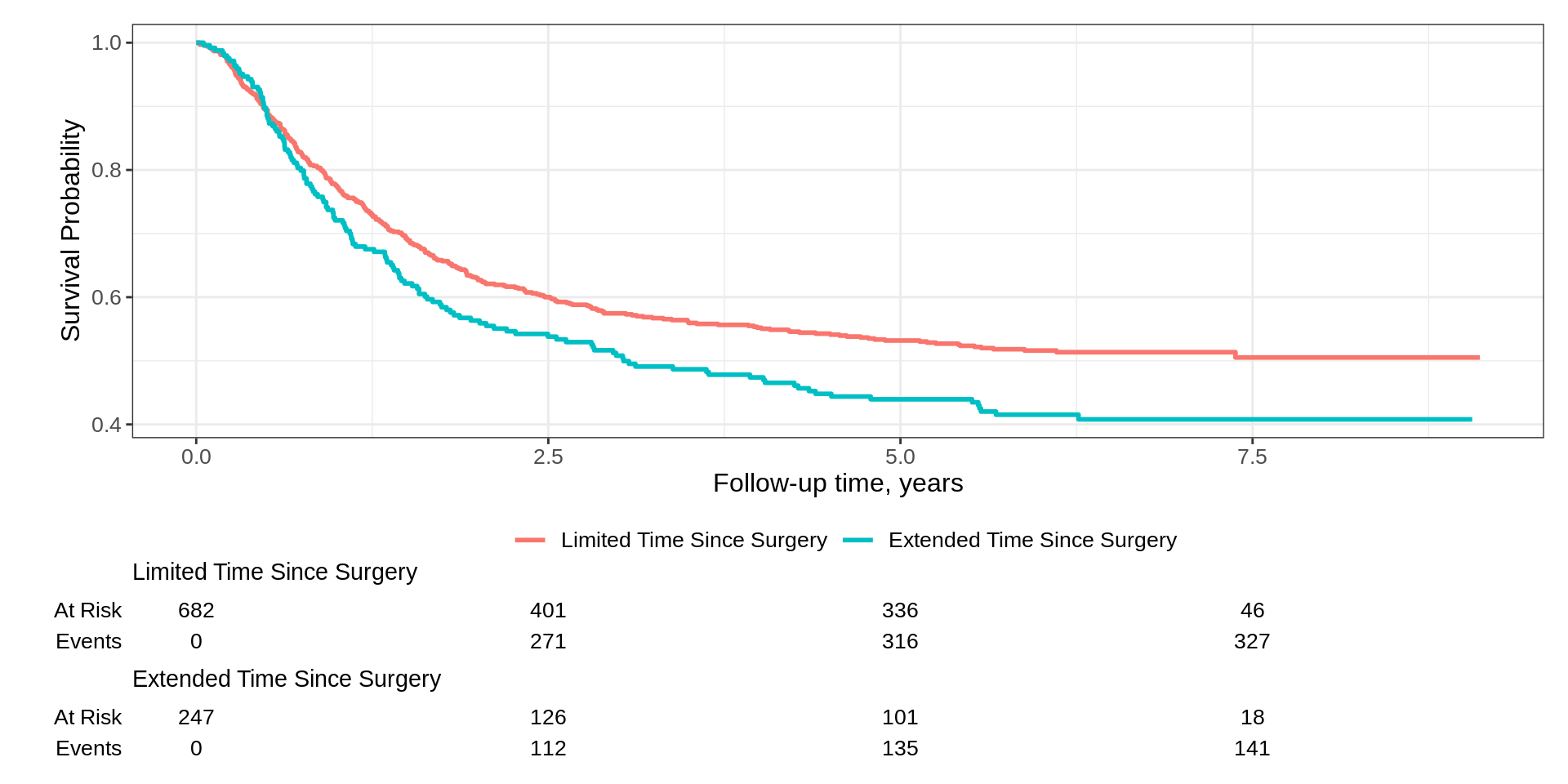
Basic Example
Padding has been reduced and curves begin in the upper left corner of plot
x-axis reports additional time points (and as a result, the risk table as well)
We updated the y-axis label weaving standard ggplot2 functions
Basic Example
Padding has been reduced and curves begin in the upper left corner of plot
x-axis reports additional time points (and as a result, the risk table as well)
We updated the y-axis label weaving standard ggplot2 functions
We can even use ggplot2-extender functions
Basic Example, transformation
Basic Example
- What is
scale_ggsurvfit()?
scale_y_continuous(expand = c(0.025, 0), limits = c(0, 1), label = scales::label_percent())
scale_x_continuous(expand = c(0.015, 0), n.breaks = 8)If you use this function, you must include all scale specifications that would appear in
scale_x_continuous()orscale_y_continuous()Do not call
scale_x_continuous()orscale_y_continuous()along withscale_ggsurvfit(): scales are not additive, rather they replace existing scales.For example, it’s common you’ll need to specify the x-axis break points.
scale_ggsurvfit(x_scales=list(breaks=0:9))
Additional examples
{ggsurvfit} defaults

{ggplot2} styled
gg_styled <-
gg_default +
coord_cartesian(xlim = c(0, 8)) +
scale_y_continuous(
limits = c(0, 1),
labels = scales::percent,
expand = c(0.01, 0)
) +
scale_x_continuous(breaks = 0:9, expand = c(0.02, 0)) +
scale_color_manual(values = c('#54738E', '#82AC7C')) +
scale_fill_manual(values = c('#54738E', '#82AC7C')) +
theme_minimal() +
theme(legend.position = "bottom") +
guides(color = guide_legend(ncol = 1)) +
labs(
title = "{ggplot2} styled",
y = "Percentage Survival"
)
gg_styled{ggplot2} styled

Risk tables
{ggsurvfit} defaults
Group by statistic or strata
Color encoding strata

Customizing the risktable statistics

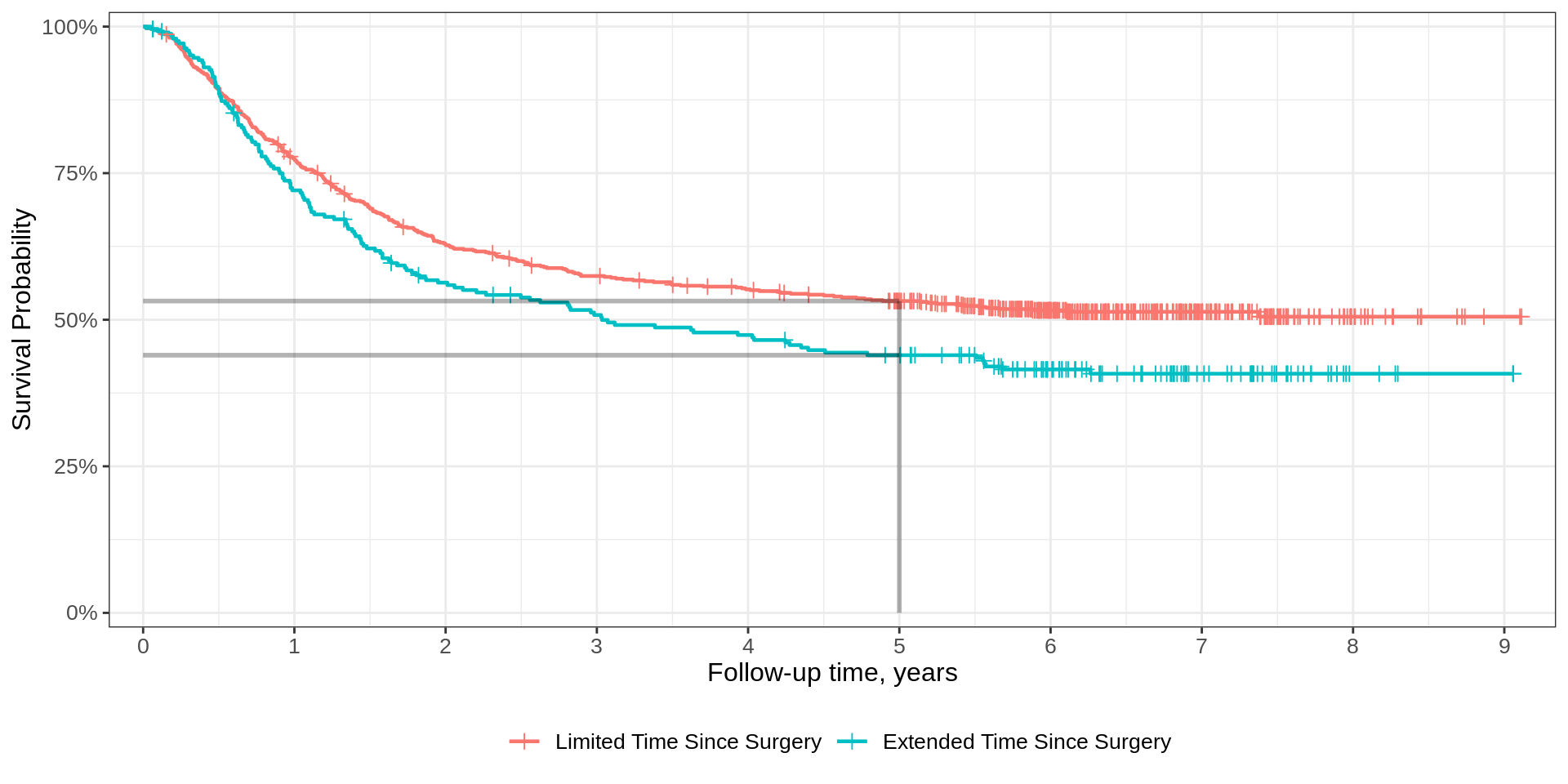
Quantiles
Median summary
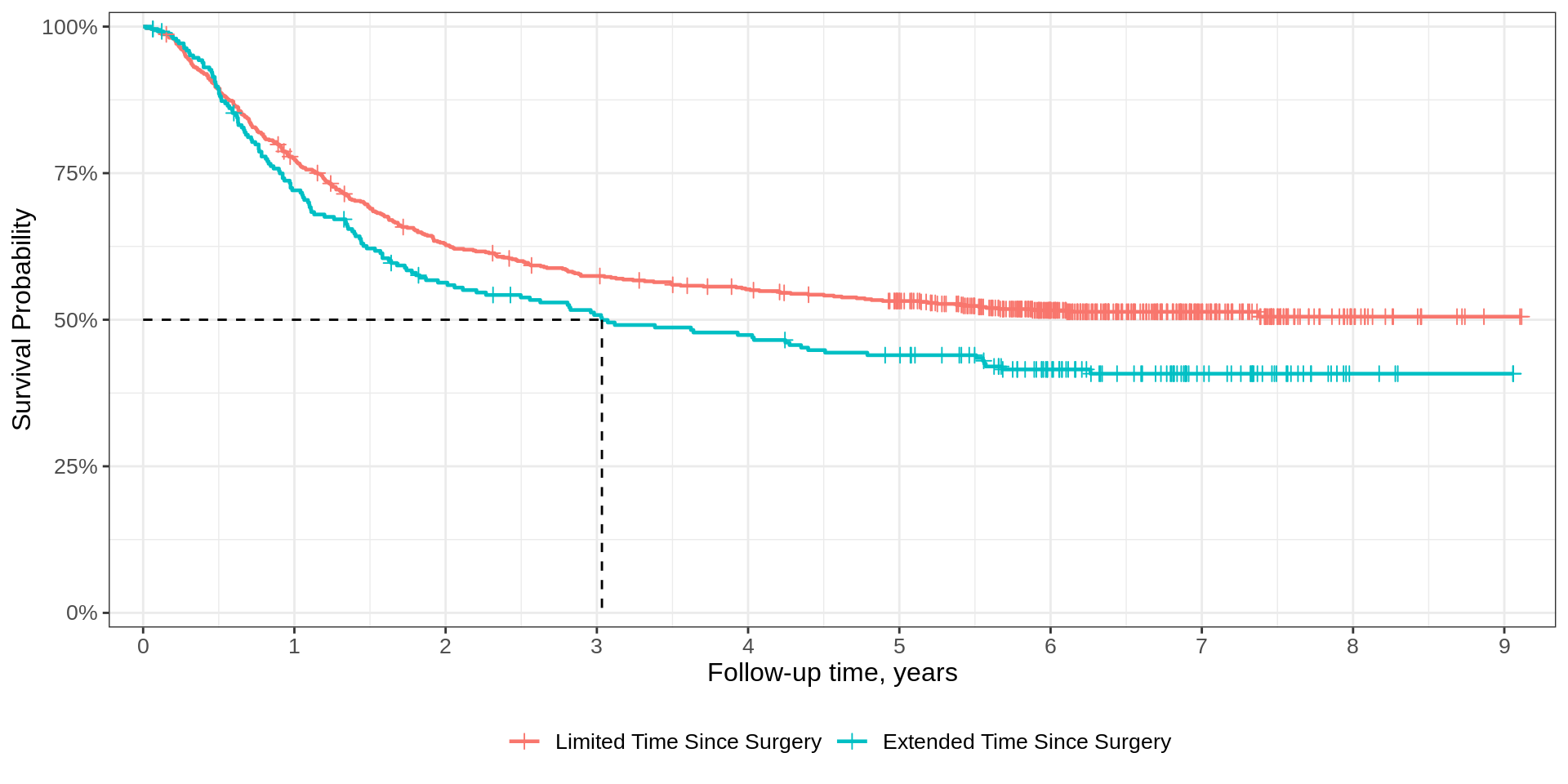
At a given timepoint
Underyling {ggsurvfit} functions
In addition to using additional {ggplot2} functions, it is helpful to understand which underlying functions are used to create the figures.
Additional arguments can be passed to the underlying functions.
| {ggsurvfit} | Underlying {ggplot2} |
|---|---|
|
|
|
|
|
|
|
|
Further Risktable Customization
Here, we customize the risktable portion of the plot.
We can style the plot area by passing ggplot2 functions to
add_risktable(theme)argument.
survfit2(Surv(time, status) ~ surg, data = df_colon) %>%
ggsurvfit(linewidth = 1) +
add_risktable(
risktable_height = 0.33,
size = 4, # increase font size of risk table statistics
theme = # increase font size of risk table title and y-axis label
list(
theme_risktable_default(axis.text.y.size = 11,
plot.title.size = 11),
theme(plot.title = element_text(face = "bold"))
)
)Further Risktable Customization

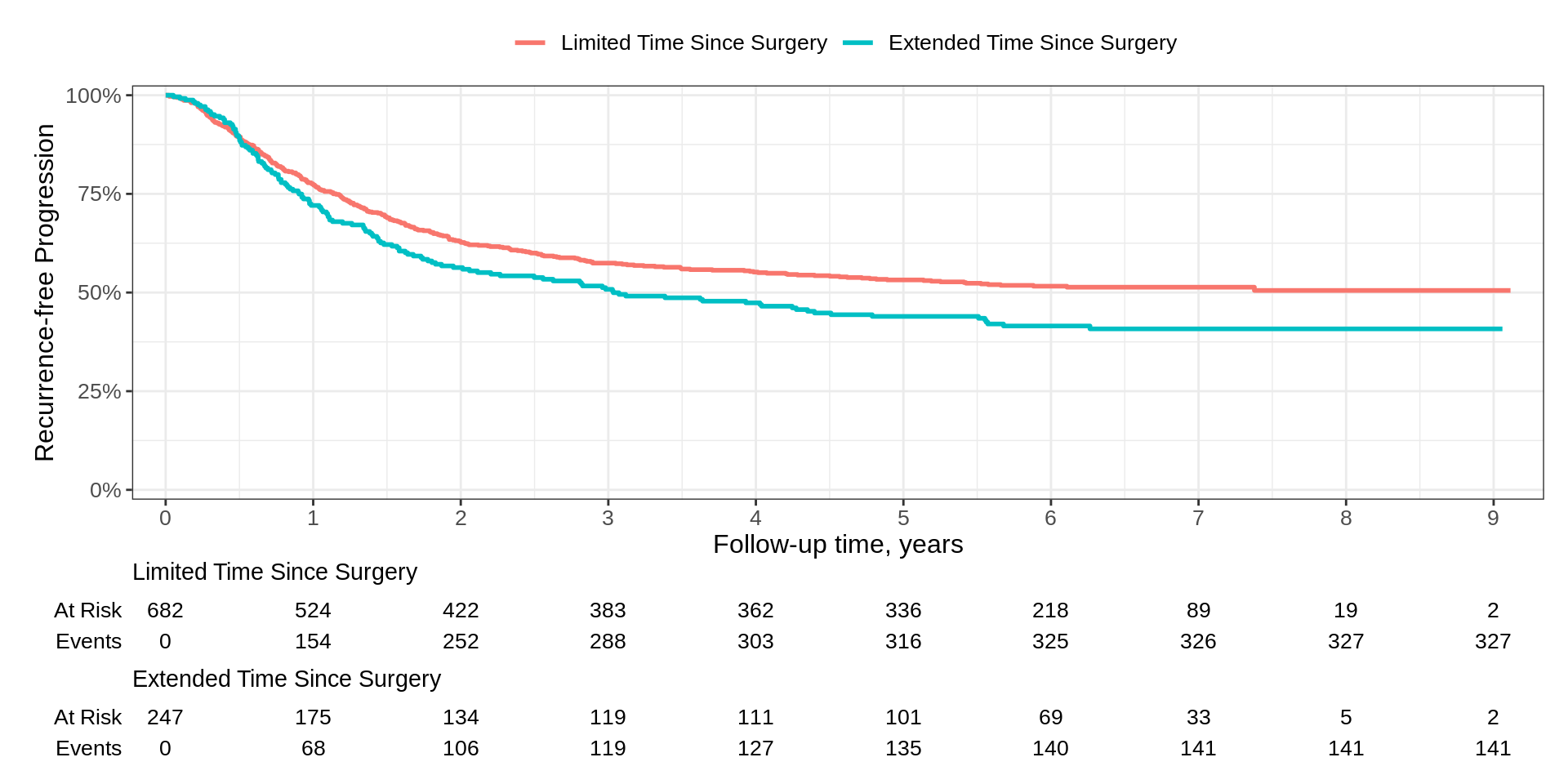
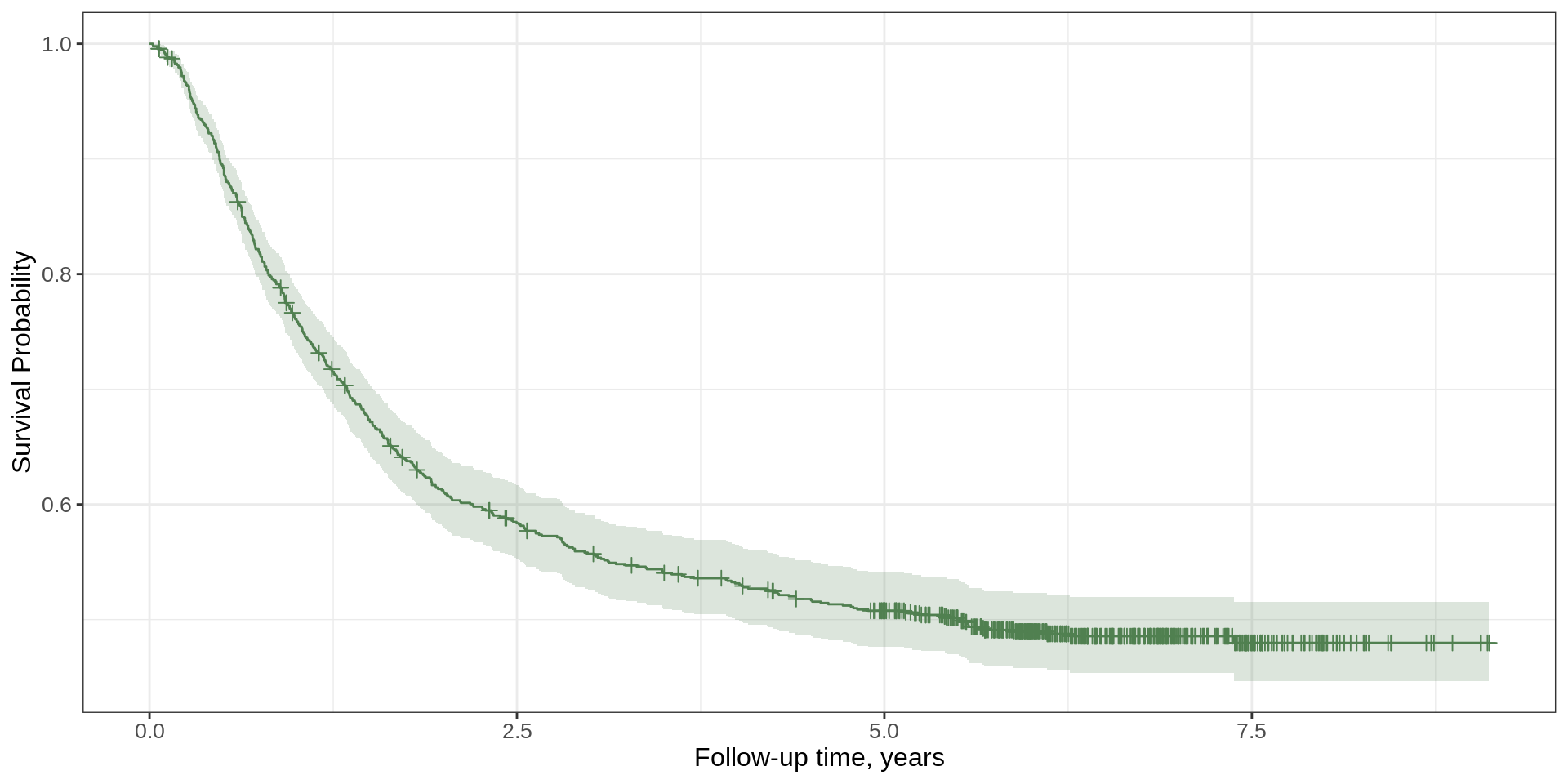
Another Example
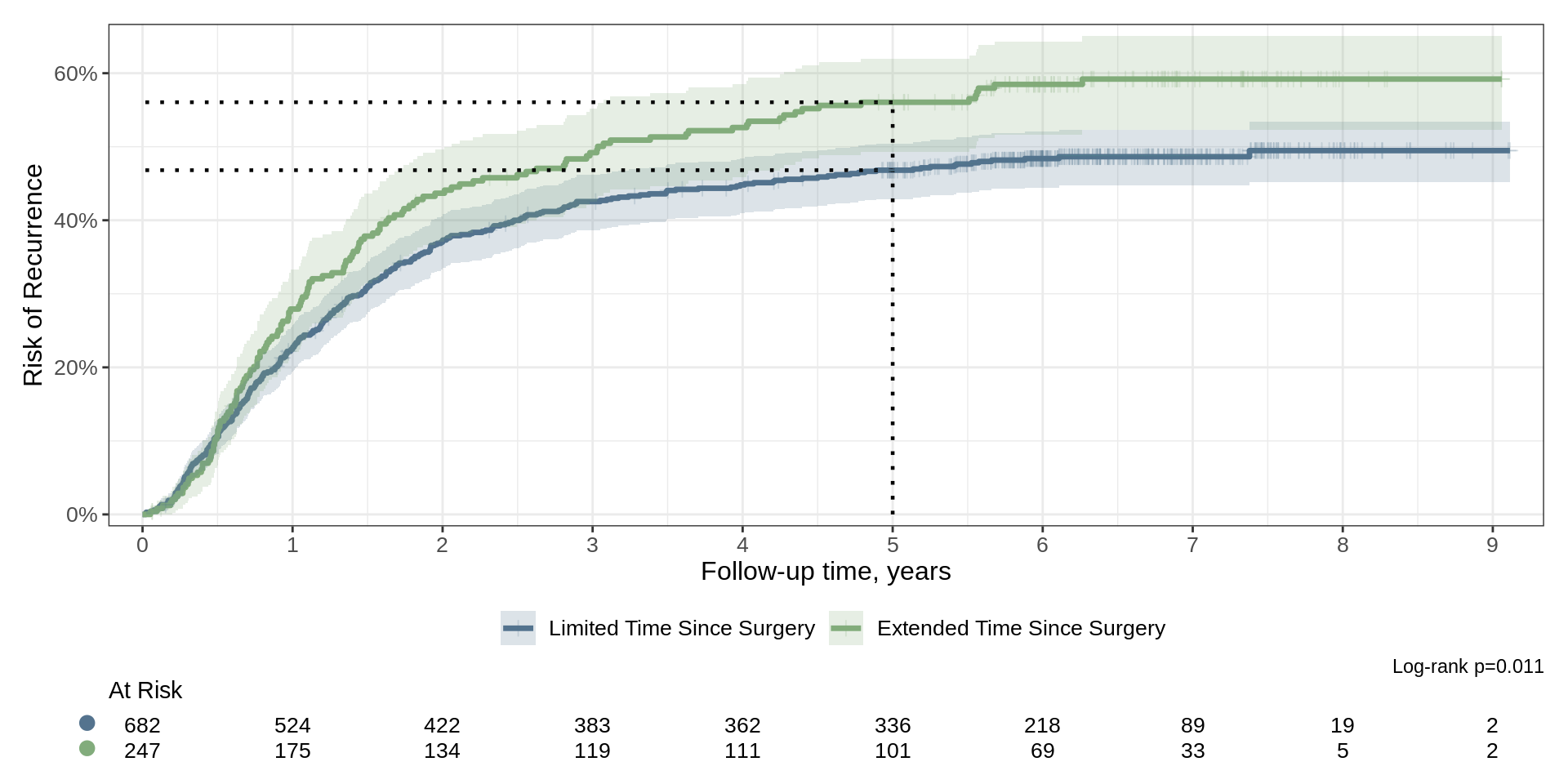
survfit2(Surv(time, status) ~ surg, data = df_colon) |>
ggsurvfit(type = "risk", linewidth = 1.2) +
add_confidence_interval() +
add_risktable(risktable_stats = "n.risk") +
add_risktable_strata_symbol(symbol = "\U25CF", size = 17) +
add_quantile(x_value = 5, linetype = "dotted", linewidth = 0.8) +
add_censor_mark(size = 2, alpha = 0.2) +
add_pvalue(caption = "Log-rank {p.value}") +
scale_ggsurvfit() +
scale_color_manual(values = c('#54738E', '#82AC7C')) +
scale_fill_manual(values = c('#54738E', '#82AC7C')) +
labs(y = "Risk of Recurrence")Another Example
KMunicate and themes
What are the elements of an effective and publishable KM plot?
There are many options to consider and many guidances available:
Morris et al. 2018 provide useful guidance for publication figures
To get figures that align with KMunicate use the
theme_ggsurvfit_KMunicate()theme along with these function options.
A note of caution on standards:
Design for your purpose, one size does not fit all
Designing means you need to think carefully about your audience and aims
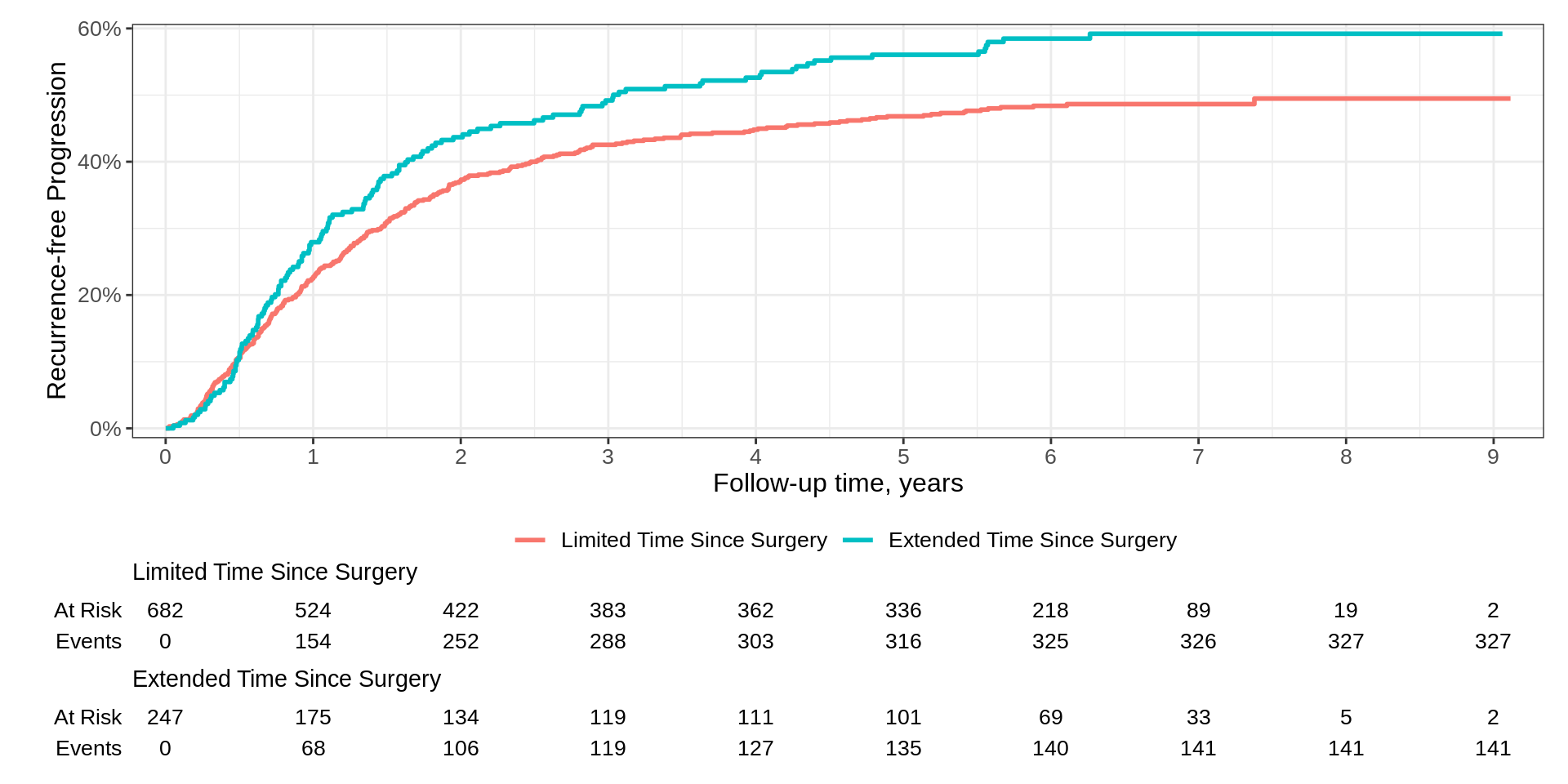
KMunicate
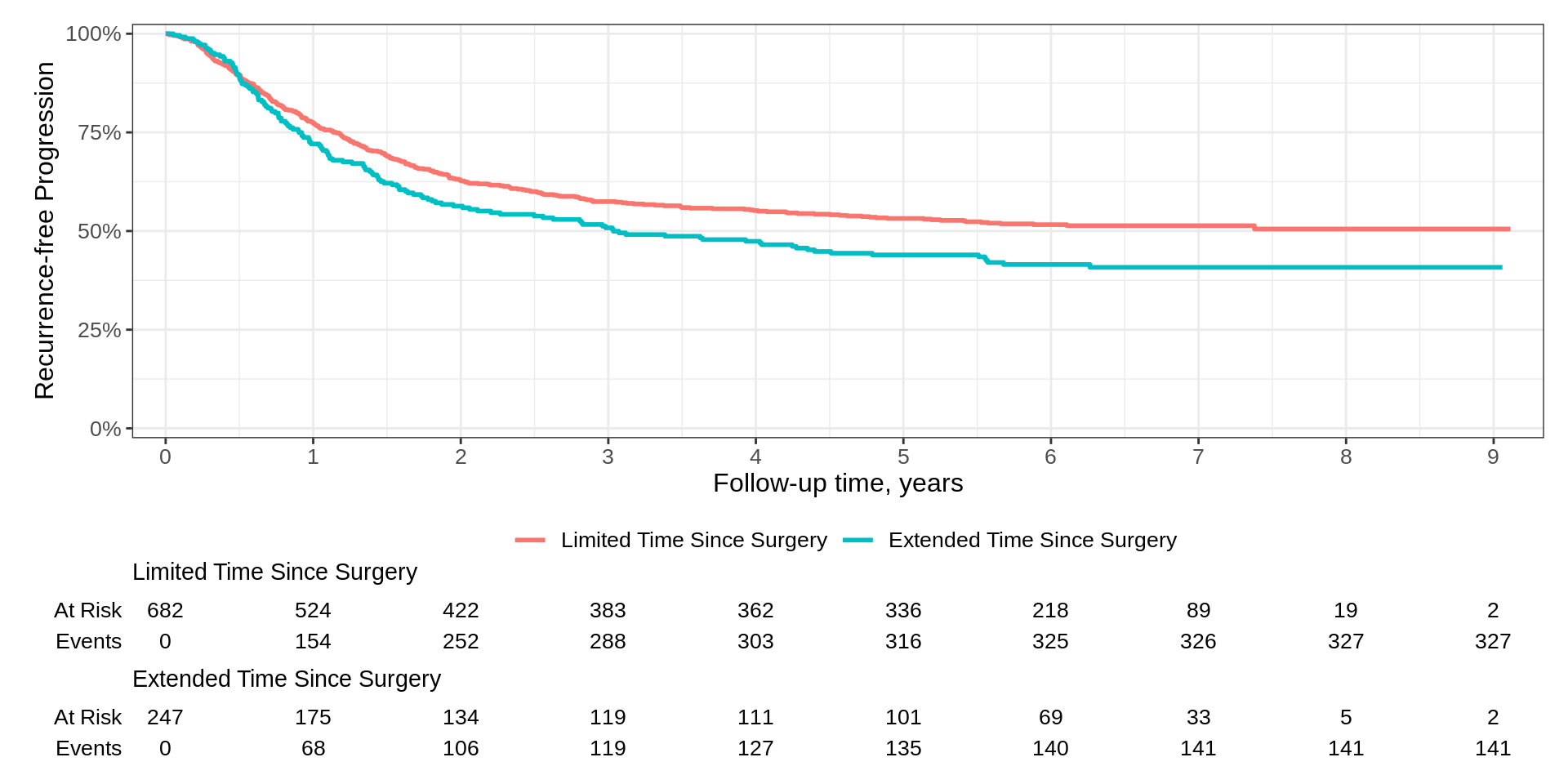
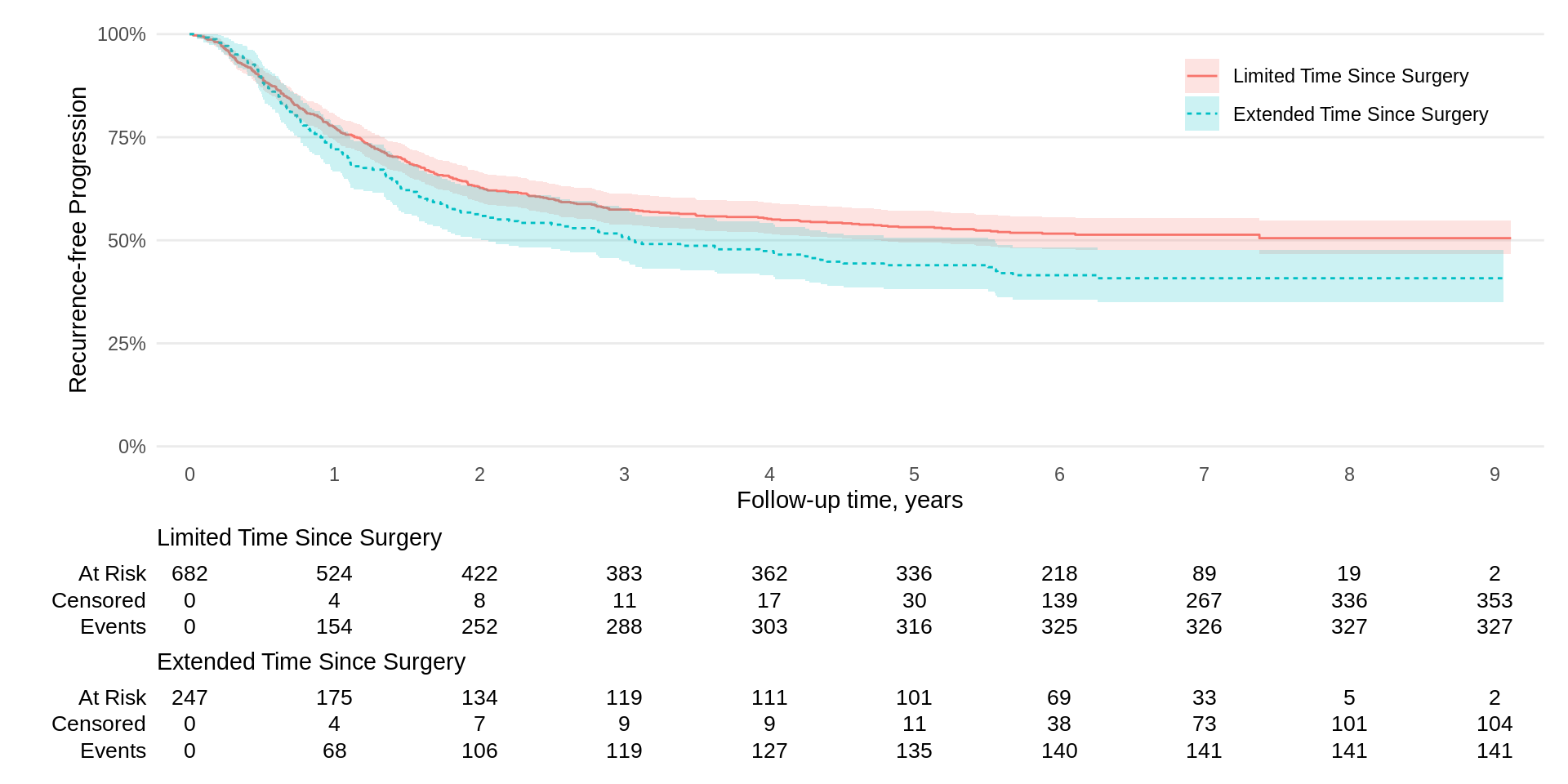
survfit2(Surv(time, status) ~ surg, data = df_colon) |>
ggsurvfit(linetype_aes = TRUE) +
add_confidence_interval() +
add_risktable(risktable_stats = c("n.risk", "cum.censor", "cum.event")) +
theme_ggsurvfit_KMunicate() +
scale_ggsurvfit() +
theme(legend.position = c(0.85, 0.85)) +
labs(y = "Recurrence-free Progression") KMunicate
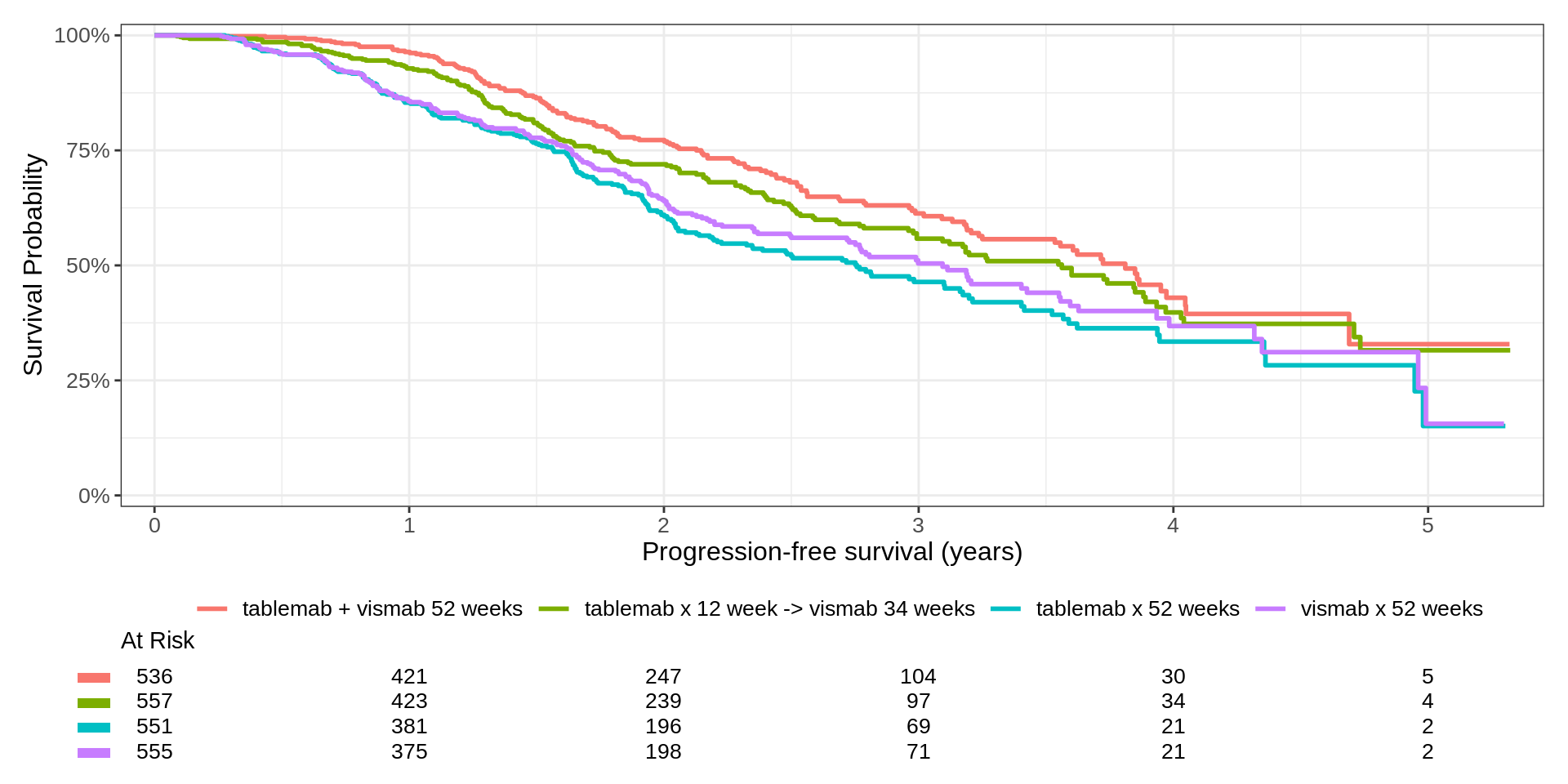
CDISC ADaM ADTTE
Gems for those using the CDISC ADaM ADTTE model.
# A tibble: 3 × 4
PARAM AVAL CNSR TRT01P
<chr> <dbl> <dbl> <chr>
1 Progression-free survival (years) 0.824 0 vismab x 52 weeks
2 Progression-free survival (years) 3.03 1 tablemab x 12 week -> vismab 34…
3 Progression-free survival (years) 2.32 0 tablemab + vismab 52 weeks - The outcome is coded in the OPPOSITE way we expect!😱😱
- The
Surv_CNSR()function handles the transformation for us survival::Surv(time = AVAL, event = 1 - CNSR, type = "right", origin = 0)- This function can be used anywhere: use it! Don’t screw it up!
- The
- The “PARAM” value is used to construct enhanced labels in the figure.
CDISC ADaM ADTTE
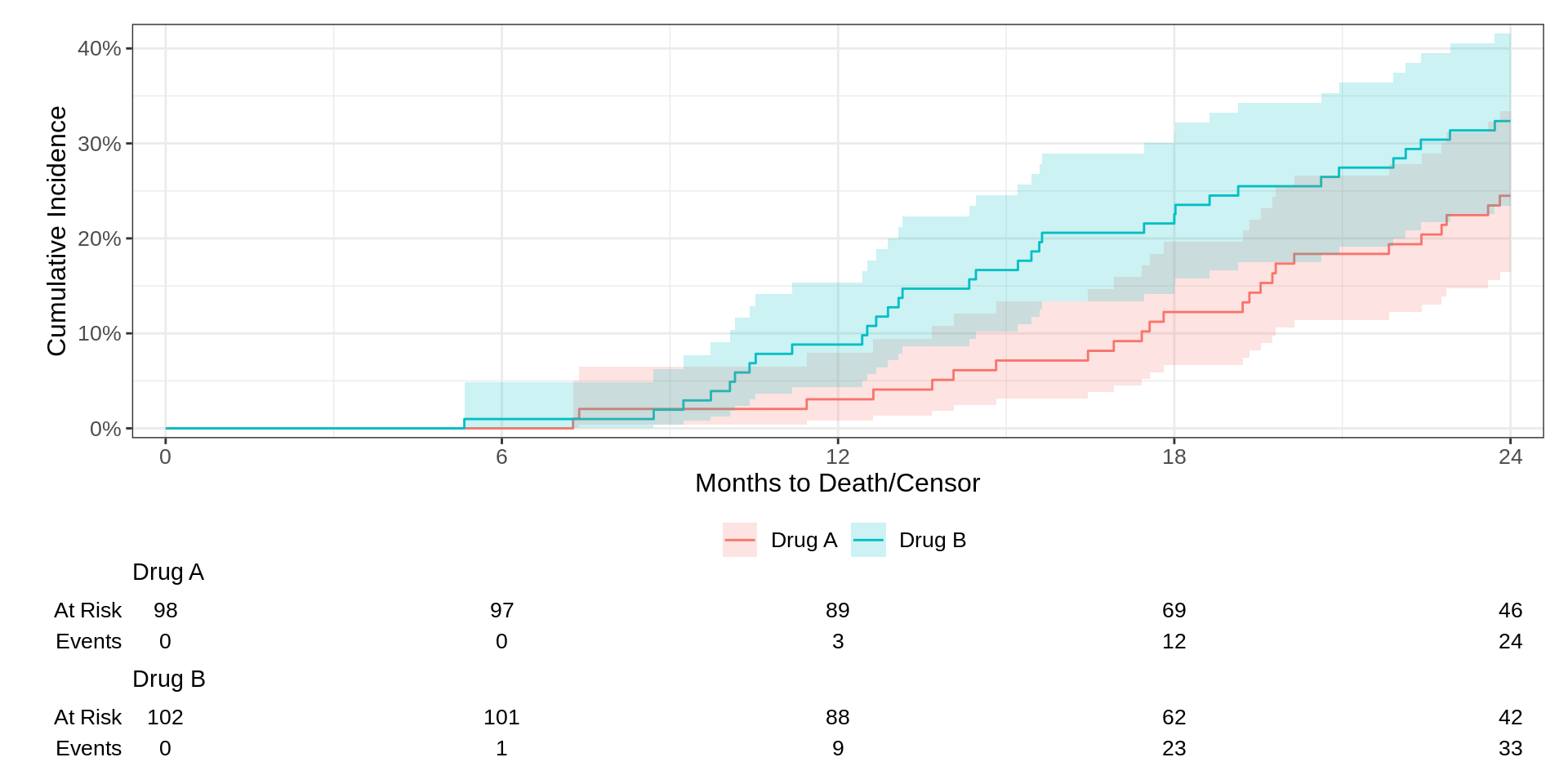
Competing Risks
There is another flavor of survival data referred to as competing risks data.
The most common type of a competing risk is an event that drives the probability of observing our event of interest to zero.
For example, if we are interested in model the time from a treatment to a disease’s return or recurrence…what do we do if a patient passes away from an unrelated cause?
In this case, death from other causes is a competing event.
The {ggsurvfit} package plays nicely with the {tidycmprsk} package for competing risks analysis.
Competing Risks
Side-by-Side
You’ll often need to place two or more figures side-by-side
My favorite package to do this is {patchwork}
But before you can use {patchwork} with {ggsurvfit}, you need to understand a bit about how these figures are constructed
The risktables are only added to the figure during printing by calling the
ggsurvfit_build()functionThis means that you must build the plot before you can cobble figures together
I have an open issue in {patchwork} to add a feature so we can avoid this extra step
Side-by-Side
p <- survfit2(Surv(time, status) ~ 1, df_colon) %>%
ggsurvfit() +
add_confidence_interval() +
add_risktable() +
scale_ggsurvfit()
# build plot (which constructs the risktable)
built_p <- ggsurvfit_build(p) |> patchwork::wrap_plots()
# I am hoping in the future was can just call `p | p`
built_p | built_p 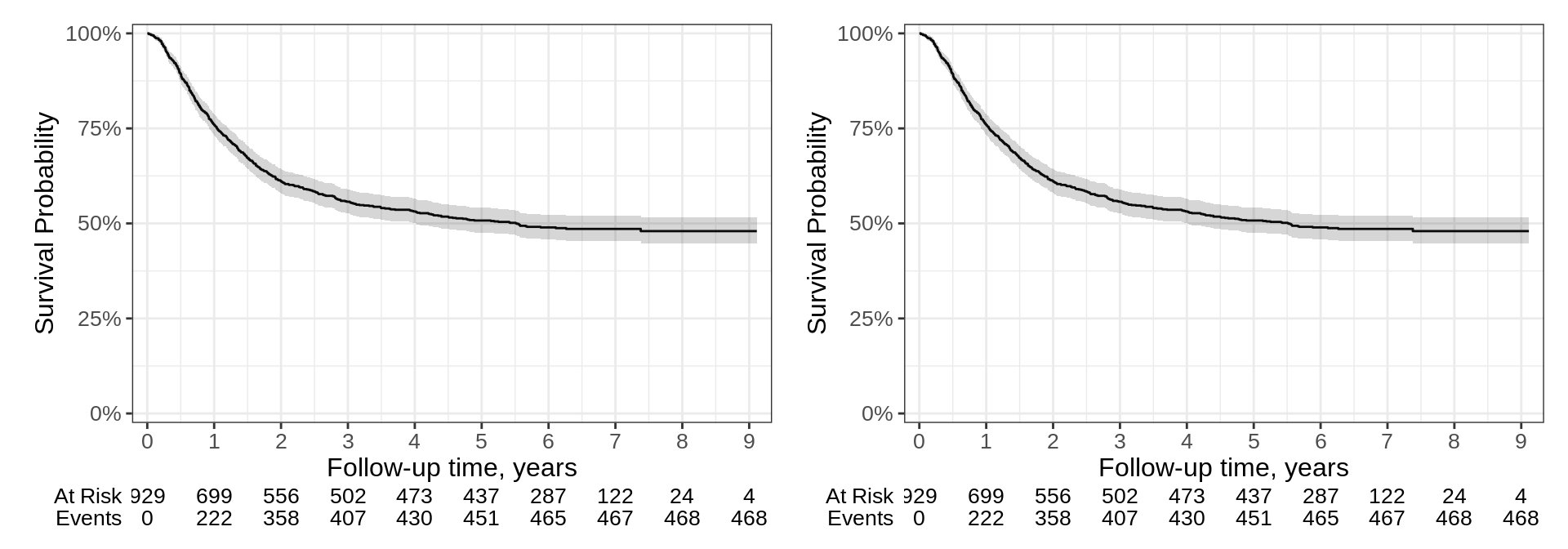
{ggsurvfit} wrap up
Ease the creation of time-to-event summary figures with ggplot2
Concise and modular code
Ready for publication or sharing figures
Sensible defaults
Also supports competing risks cumulative incidence summaries