Very experimental and early stage package for creating forest plots from gtsummary tables. The result is a gt table that includes the plot.
Installation
You can install the development version of gtforester from GitHub with:
# install.packages("devtools")
devtools::install_github("ddsjoberg/gtforester")Example
library(gtforester)
#> Loading required package: gtsummary
tbl <-
trial %>%
select(age, marker, grade, response) %>%
tbl_uvregression(
y = response,
method = glm,
method.args = list(family = binomial),
exponentiate = TRUE,
hide_n = TRUE
) %>%
modify_column_merge(
pattern = "{estimate} (95% CI {ci}; {p.value})",
rows = !is.na(estimate)
) %>%
modify_header(estimate = "**Odds Ratio**") %>%
bold_labels() %>%
add_forest()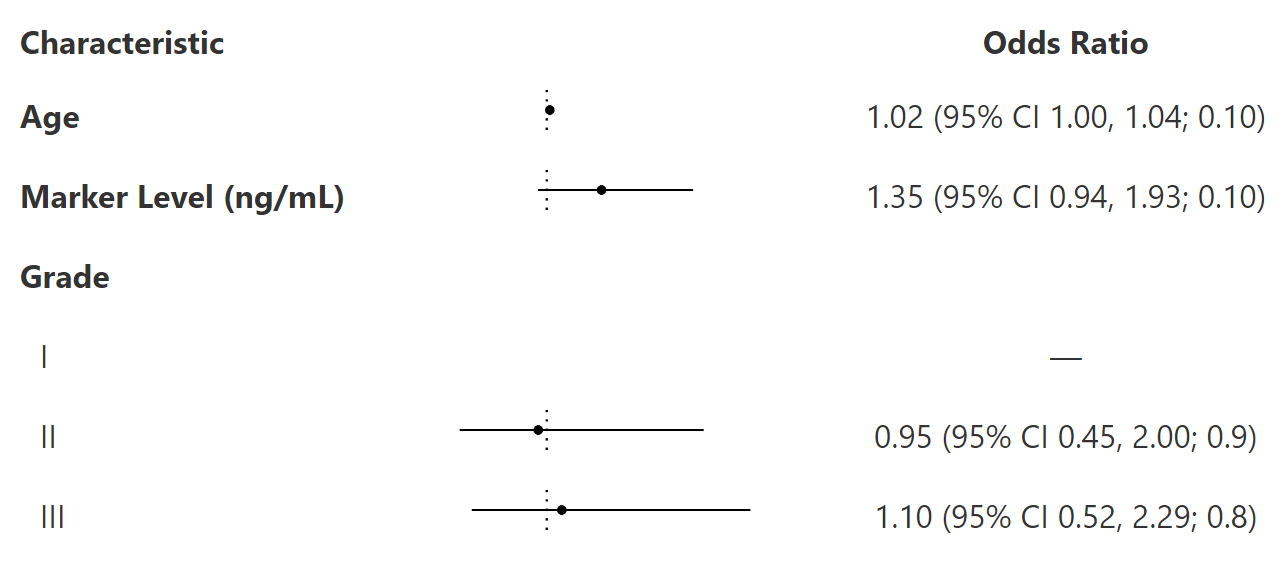
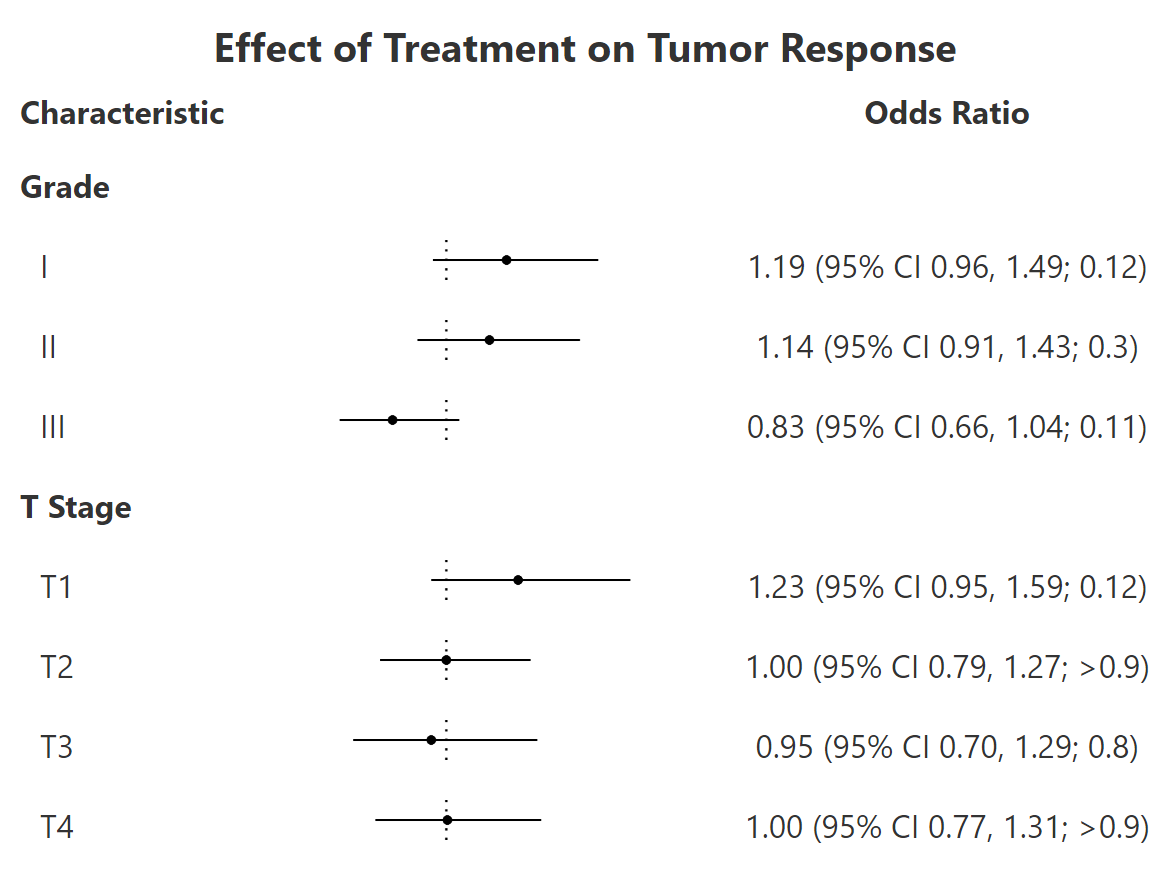
tbl <-
trial %>%
tbl_subgroups(
subgroups = c("grade", "stage"),
~ glm(response ~ trt, data = .x) %>%
gtsummary::tbl_regression(
show_single_row = trt,
exponentiate = TRUE)
) %>%
gtsummary::modify_column_merge(
pattern = "{estimate} (95% CI {ci}; {p.value})",
rows = !is.na(estimate)
) %>%
gtsummary::modify_header(estimate = "**Odds Ratio**") %>%
gtsummary::bold_labels() %>%
add_forest() %>%
gt::tab_header(gt::md("**Effect of Treatment on Tumor Response**"))