The aim of Surv_CNSR() is to map the inconsistency in data convention between
the survival package and
CDISC ADaM ADTTE data model.
The function creates a survival object (e.g. survival::Surv()) that
uses CDISC ADaM ADTTE coding conventions and converts the arguments to the
status/event variable convention used in the
survival package.
The AVAL and CNSR arguments are passed to
survival::Surv(time = AVAL, event = 1 - CNSR, type = "right", origin = 0).
Details
The Surv_CNSR() function creates a survival object utilizing the
expected data structure in the CDISC ADaM ADTTE data model,
mapping the CDISC ADaM ADTTE coding conventions with the expected
status/event variable convention used in the survival package—specifically,
the coding convention used for the status/event indicator.
The survival package expects the status/event indicator in the
following format: 0=alive, 1=dead. Other accepted choices are
TRUE/FALSE (TRUE = death) or 1/2 (2=death).
A final but risky option is to omit the indicator variable, in which case
all subjects are assumed to have an event.
The CDISC ADaM ADTTE data model adopts a different coding convention for
the event/status indicator. Using this convention, the event/status variable
is named 'CNSR' and uses the following coding: censor = 1, status/event = 0.
Examples
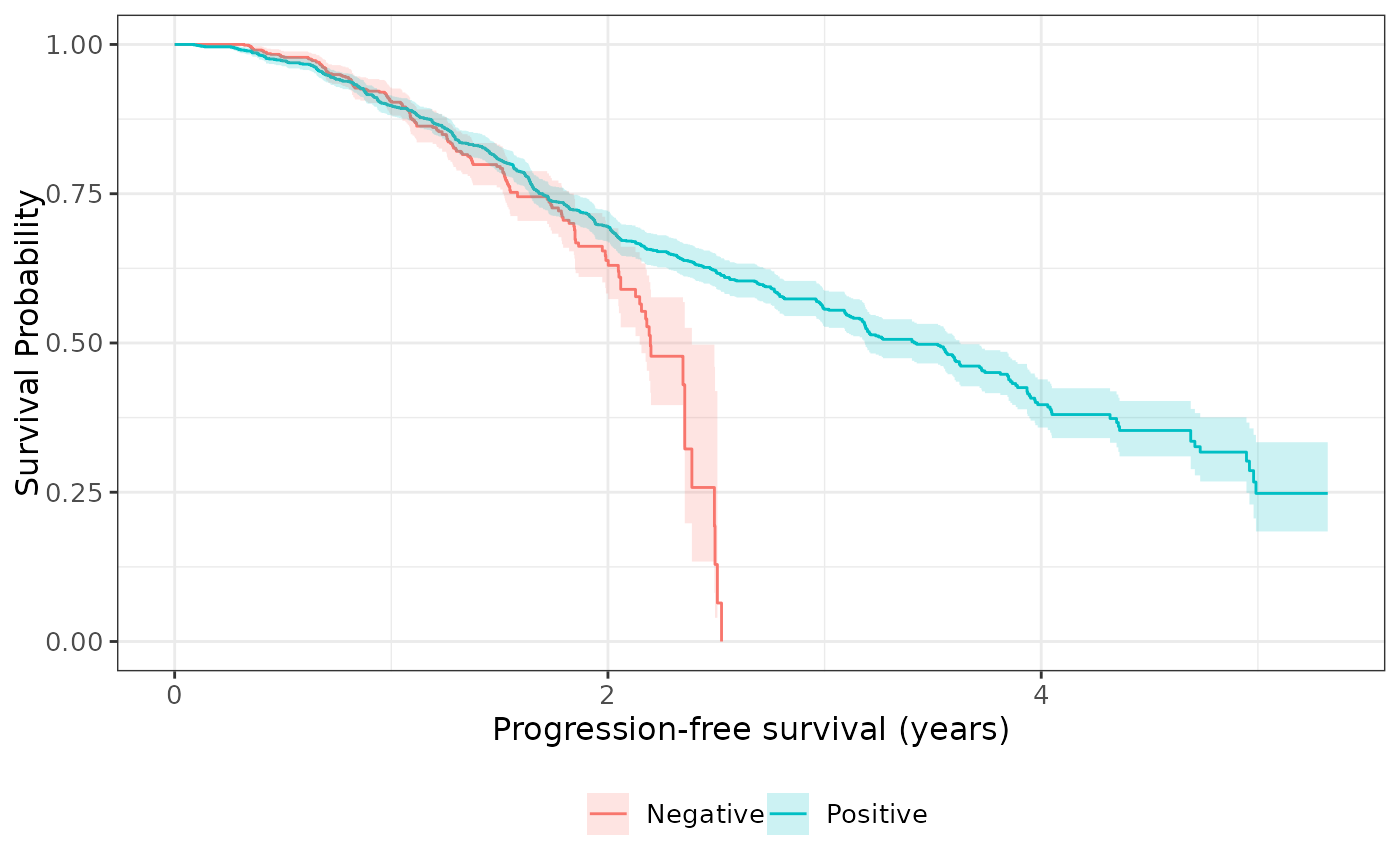
# Use the `Surv_CNSR()` function with ggsurvfit functions
survfit2(formula = Surv_CNSR() ~ STR01, data = adtte) %>%
ggsurvfit() +
add_confidence_interval()
 # Use the `Surv_CNSR()` function with functions from other packages as well
survival::survfit(Surv_CNSR() ~ STR01, data = adtte)
#> Call: survfit(formula = Surv_CNSR() ~ STR01, data = adtte)
#>
#> n events median 0.95LCL 0.95UCL
#> STR01=Negative 893 166 2.20 2.15 2.39
#> STR01=Positive 1306 589 3.41 3.19 3.63
survival::survreg(Surv_CNSR() ~ STR01 + AGE, data = adtte) %>%
broom::tidy()
#> # A tibble: 4 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 0.943 0.147 6.40 1.56e-10
#> 2 STR01Positive 0.213 0.0534 3.98 6.78e- 5
#> 3 AGE 0.00376 0.00221 1.70 8.97e- 2
#> 4 Log(scale) -0.543 0.0306 -17.8 1.47e-70
# Use the `Surv_CNSR()` function with functions from other packages as well
survival::survfit(Surv_CNSR() ~ STR01, data = adtte)
#> Call: survfit(formula = Surv_CNSR() ~ STR01, data = adtte)
#>
#> n events median 0.95LCL 0.95UCL
#> STR01=Negative 893 166 2.20 2.15 2.39
#> STR01=Positive 1306 589 3.41 3.19 3.63
survival::survreg(Surv_CNSR() ~ STR01 + AGE, data = adtte) %>%
broom::tidy()
#> # A tibble: 4 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 0.943 0.147 6.40 1.56e-10
#> 2 STR01Positive 0.213 0.0534 3.98 6.78e- 5
#> 3 AGE 0.00376 0.00221 1.70 8.97e- 2
#> 4 Log(scale) -0.543 0.0306 -17.8 1.47e-70
